Mustguseal
A web-server for Multiple Structure-Guided Sequence Alignment of Protein Families
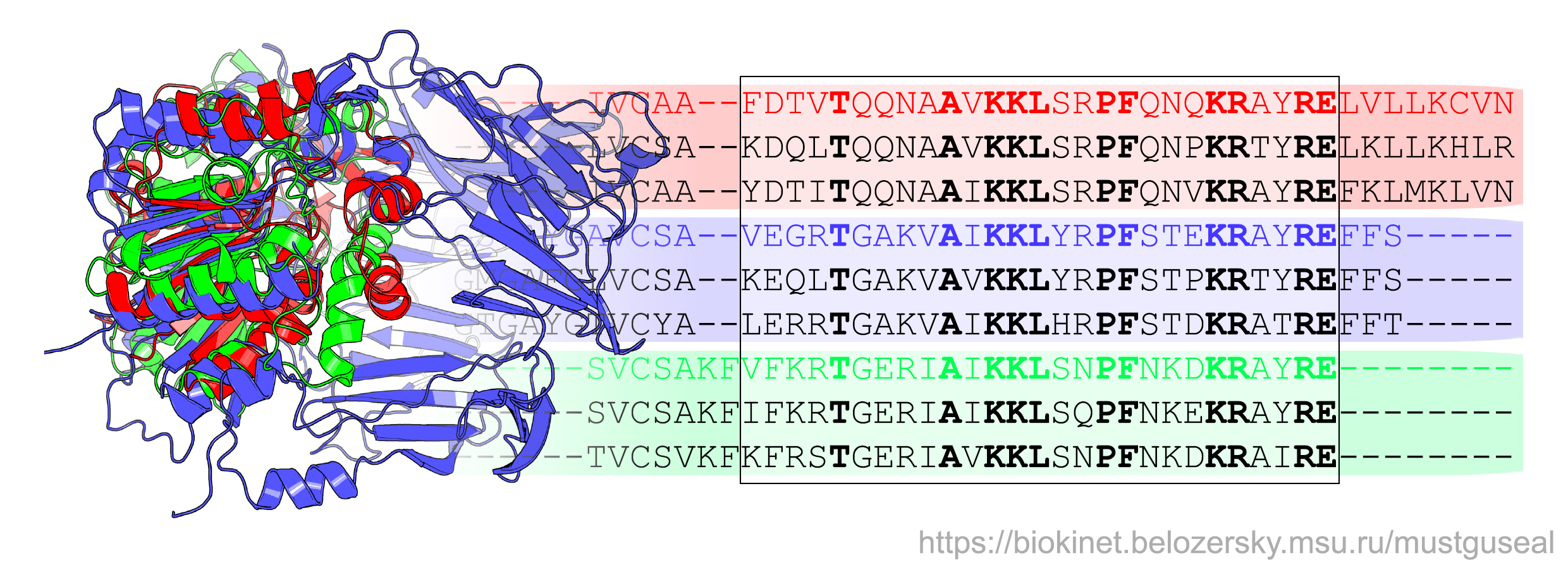
Mustguseal can automatically construct large structure-based sequence alignments of functionally diverse protein families that include thousands of proteins based on all available information about their structures and sequences in public databases
to help at studying a protein function and regulation, designing improved enzyme variants for practical application and selective modulators of enzyme functional properties
Alternatively, you can use [The compact version of the input page]
Server version A 1.6 since Jan 13th, 2021 Click here to view version history
THIS WEB-SITE/WEB-SERVER/SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS OR HARDWARE OWNERS OR WEB-SITE/WEB-SERVER/SOFTWARE MAINTEINERS/ADMINISTRATORS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE WEB-SITE/WEB-SERVER/SOFTWARE OR THE USE OR OTHER DEALINGS IN THE WEB-SITE/WEB-SERVER/SOFTWARE. PLEASE NOTE THAT OUR WEBSITE COLLECTS STANDARD APACHE2 LOGS, INCLUDING DATES, TIMES, IP ADDRESSES, AND SPECIFIC WEB ADDRESSES ACCESSED. THIS DATA IS STORED FOR APPROXIMATELY ONE YEAR FOR SECURITY AND ANALYTICAL PURPOSES. YOUR PRIVACY IS IMPORTANT TO US, AND WE USE THIS INFORMATION SOLELY FOR WEBSITE IMPROVEMENT AND PROTECTION AGAINST POTENTIAL THREATS. BY USING OUR SITE, YOU CONSENT TO THIS DATA COLLECTION.
Want to understand how Mustguseal works?
Step 1: Press the green button "Mustguseal it NOW!" above
Step 2: Scroll down on that page, check out the "Algorithm outline" on the right side
Step 3: Press the blue button "Demo mode" on that page
Practical guide: Suplatov D., Sharapova Y., Švedas V. (2021) Mustguseal and Sister Web-Methods: A Practical Guide to Bioinformatic Analysis of Protein Superfamilies. In: Katoh K. (eds) Multiple Sequence Alignment. Methods in Molecular Biology, vol 2231. Humana, New York, NY. DOI: 10.1007/978-1-0716-1036-7_12.
Protocol: Suplatov D.A., Kopylov K.E., Popova N.N., Voevodin V.V., Švedas V.K. (2018) Mustguseal: a Server for Multiple Structure-Guided Sequence Alignment of Protein Families. Bioinformatics, 34(9), 1583-1585. DOI: 10.1093/bioinformatics/btx831. Supplementary Data are available from the authors or at Bioinformatics online
Conference: D. Suplatov, Y. Sharapova, D. Timonina, E. Schmalhausen, K. Fesko, N. Popova, V. Muronets, V. Voevodin, V. Švedas (2019) Openaccess Mustguseal platform for bioinformatic analysis in computational enzymology. FEBS Congress 2019, poster: [download]
Original research carried out with the help of Mustguseal:
Mustguseal was used to study domain organisation of modular neuraminidases/sialidases Sharapova Y.A. et al. (2018) FEBS J. DOI:10.1111/febs.14486
Mustguseal was used to study sequence/structure/function relationship, reaction specificity determinants, and promiscuous catalytic activity in PLP fold-type I dependent enzymes Fesko K. et al. (2018) FEBS Open Bio. DOI:10.1002/2211-5463.12441
Mustguseal was used to study binding specificity of allosteric inhibitor Doramapimod to human p38alpha MAP kinase and related Serine/Threonine-kinases Suplatov D. et al. (2018) J. Biomol. Struct. Dyn. DOI:10.1080/07391102.2018.1475260 View the highlights for this paper on Kudos - click here.
Mustguseal was used to study the structural organization of a novel PLP fold-type IV dependent transaminase from Thermobacullum terrenum featuring a unique substrate specificity Bezsudnova E. et al. (2019) Biochimie. DOI:10.1016/j.biochi.2018.12.017
Site Navigation:
- Presentation of the Mustguseal Protocol and Server
- The Mustugeal Protocol in a nutshell
- Overview of the Input modes
- Input Parameters and their influence over the results
- The Output: Results and Analysis
- Mustguseal Performance
- Mustguseal File Sharing and Security Features
- Post processing of a very large Mustguseal alignment using MAPU
- Advanced tools to further study the Mustguseal alignment
- Citing Mustguseal Protocol and Server
The Mustguseal FAQs:
- What is Mustguseal?
- How Mustguseal works?
- What software is required to use Mustguseal?
- What web-browser is required to use Mustguseal?
- How to cite Mustguseal?
- How can a Mustguseal alignment help in your research?
What is Mustguseal? Mustguseal stands for Multiple Structure-Guided Sequence Alignment. Mustguseal is a bioinformatic protocol designed to build large alignments of functionally diverse protein families and a web-platform to provide a user-friendly web-based interface to the Mustguseal protocol through the World Wide Web. Mustguseal can be used to build focused alignments of the selected protein families or superimpose a large collection of related proteins within a superfamily. This server is capable of constructing multiple alignments of functionally diverse families that include thousands of proteins basing on all available information about their structures and sequences in public databases. In addition, three integrated web-servers are available to further study the obtained alignments. You should see the Presentation of the Mustguseal server for more information.
How Mustguseal works? The Mustguseal protocol implements structure similarity search to collect evolutionarily remote relatives, which are expected to represent different protein families. Then, for each collected evolutionarily remote relative, Mustguseal runs a sequence similarity search to collect evolutionarily close relatives - members of the corresponding families. In such a way Mustguseal takes into account variability of sequences and structures within a large superfamily to obtain a set of functionally diverse homologous proteins. A combination of structure and sequence alignment procedures is then implemented to build the final multiple alignment.
What software is required to use Mustguseal? All steps of the Mustguseal protocol are executed entirely on the server side. You do not need any specific software on your side. The Analysis section of the Results page offers interactive content for sequence and structure analysis. Interactivity is implemented in HTML5 and therefore no plugins nor Java are required. The only prerequisite for using the Analysis section of the Results is a HTML5-compatible web-browser. Current versions of all major browsers support HTML5 as the default standard.
What web-browser is required to use Mustguseal? The Mustguseal web-server is not browser-specific. You would need an HTML5-compatible web-browser to use the Analysis section of the Results. To ensure the HTML5-compatibility upgrade your current browser to the latest version. The server has been successfully tested for compatibility with the following browsers:
- Google Chrome (https://www.google.com/chrome/);
- Mozilla Firefox (https://www.mozilla.org/en-US/firefox/new);
- KDE Konqueror (https://konqueror.org/);
- Microsoft Edge.
Please note that we have had compatibility issues with the Microsoft Internet Explorer and therefore this browser is currently not supported by the Mustguseal.
How to cite Mustguseal? See this page for details.
How can a Mustguseal alignment help in your research? You should use sister web-servers of Mustguseal to further study the obtained alignments:
- the Zebra web-server to identify and prioritize conserved and specific positions in a functionally diverse superfamily and to select hot-spots for rational design of the query protein;
- the pocketZebra web-server to identify and rank binding sites in proteins by functional significance and select particular positions in the structure that are important for selective binding of substrates/inhibitors/effectors;
- the visualCMAT web-server to select and interpret correlated mutations/co-evolving residues in protein families;
- the Yosshi web-server to systematically classify and study disulfide bonds in your protein families, and select hot-spots for disulfide engineering in the structure of your query protein.
Implementation of Mustguseal, Zebra, pocketZebra, visualCMAT, and Yosshi in the laboratory practice can help at studying mechanisms of protein action, designing enzymes with improved properties for practical application and selective modulators of activity of the wild-type proteins. Find out more about the advanced tools to study the Mustguseal alignment.
