
pocketZebra
Web-server For Selection and Classification of Subfamily-Specific Binding Sites
by Bioinformatic Analysis Of Diverse Protein Families
Have a protein family? ...
... to identify and rank binding sites in proteins by functional significance
and select particular positions in the structure that are important for selective binding of substrates/inhibitors/effectors
THIS WEB-SITE/WEB-SERVER/SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS OR HARDWARE OWNERS OR WEB-SITE/WEB-SERVER/SOFTWARE MAINTEINERS/ADMINISTRATORS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE WEB-SITE/WEB-SERVER/SOFTWARE OR THE USE OR OTHER DEALINGS IN THE WEB-SITE/WEB-SERVER/SOFTWARE. PLEASE NOTE THAT OUR WEBSITE COLLECTS STANDARD APACHE2 LOGS, INCLUDING DATES, TIMES, IP ADDRESSES, AND SPECIFIC WEB ADDRESSES ACCESSED. THIS DATA IS STORED FOR APPROXIMATELY ONE YEAR FOR SECURITY AND ANALYTICAL PURPOSES. YOUR PRIVACY IS IMPORTANT TO US, AND WE USE THIS INFORMATION SOLELY FOR WEBSITE IMPROVEMENT AND PROTECTION AGAINST POTENTIAL THREATS. BY USING OUR SITE, YOU CONSENT TO THIS DATA COLLECTION.
No alignment? No problem!
Press  to run the Mustguseal web-server with a pre-selected set of parameters and automatically construct a multiple alignment of your protein families for further analysis by the pocketZebra
to run the Mustguseal web-server with a pre-selected set of parameters and automatically construct a multiple alignment of your protein families for further analysis by the pocketZebra
Navigation
- The algorithm outline
- Explanation of the input data
- Explanation of the output data
- Examples and benchmarking
Publication Suplatov D., Kirilin E., Arbatsky M., Takhaveev V., Švedas V. (2014) pocketZebra: a web-server for automated selection and classification of subfamily-specific binding sites by bioinformatic analysis of diverse protein families. Nucl. Acids Res. 42(W1): W344-W349. DOI:dx.doi.org
Acknowledgements Financial support from Skolkovo Institute of Science and Technology [grant 182-MRA]
Questions other people ask about pocketZebra:
Q: What is pocketZebra?
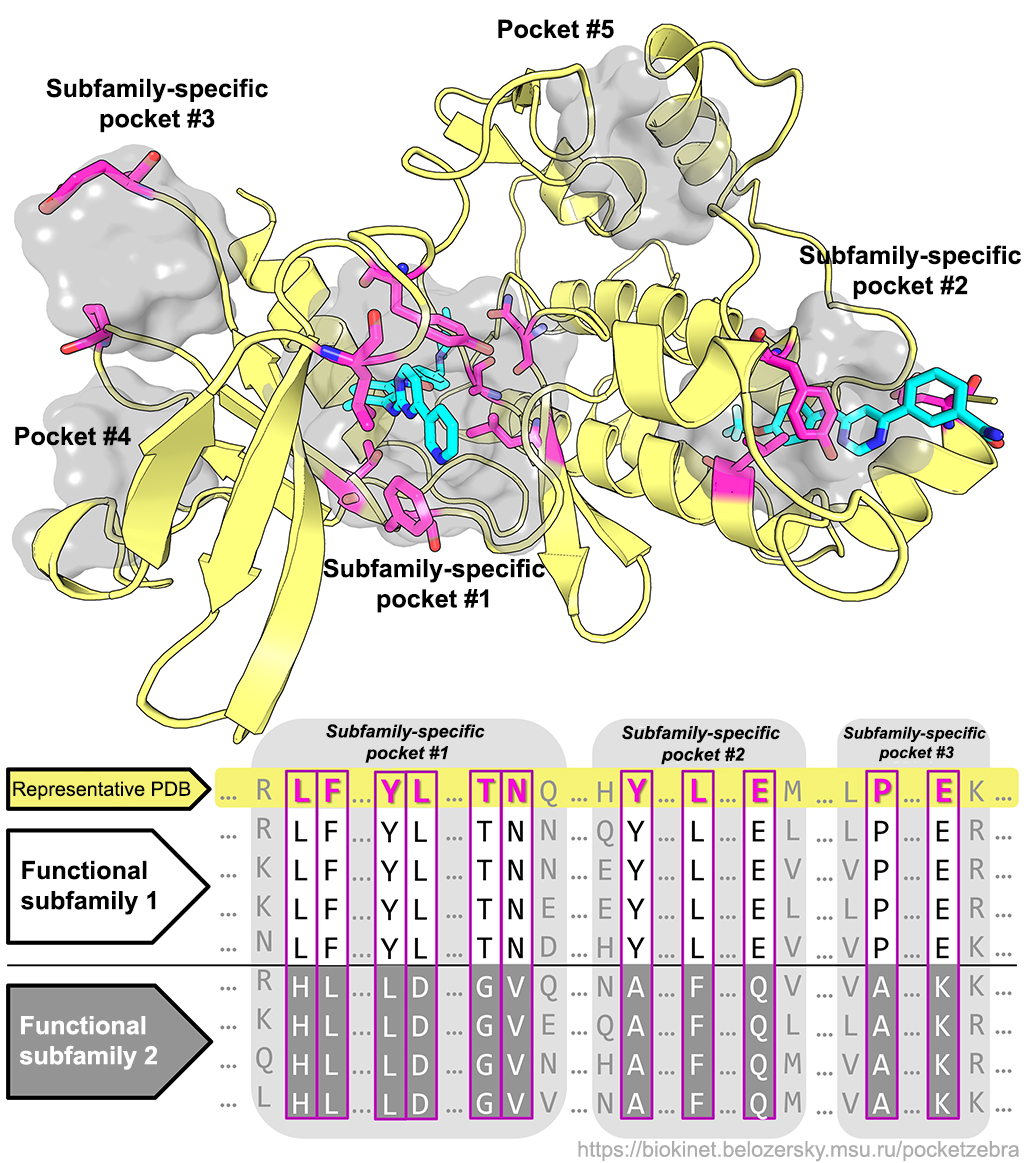
Q: Why are the subfamily-specific positions (SSPs) important?
Q: What do I need to run pocketZebra?
Q: Where can I get a multiple sequence alignment for my protein family?
Q: What do I get from pocketZebra?
Q: How can I cite pocketZebra
Q: What is a JobID?
Q: I have problems starting the interactive 3D structure viewer at the results page
Q: What is pocketZebra? A: pocketZebra is a new web-server that implements the power of bioinformatics and geometry-based structural approaches to detect and rank binding sites in proteins by their functional significance. Additionally, pocketZebra will identify a list of amino acid residues that are involved in selective binding of ligands. pocketZebra can be used to annotate functional and regulatory sites in protein families and design novel selective inhibitors/effectors. The website is free and open to all users and there is no login requirement.
Q: Why are the subfamily-specific positions (SSPs) important? A: Proteins within a single family share a common function but differ in more specific properties and can be divided into subfamilies with different specificity, enantioselectivity, stability, etc. Completely conserved positions can define general properties of the entire family (for example, have direct roles in enzyme catalytic machinery) but do not explain functional diversity. On the opposite the subfamily-specific positions (SSPs) – conserved only within protein subfamilies, but different between subfamilies – seem to be responsible for functional diversity (different substrate specificity, catalytic activity, stability, etc.) including selective binding of various substrates. SSPs can contribute to a better understanding of structure-function relationship in proteins and guide the design of novel selective inhibitors/effectors.
Q: What do I need to run pocketZebra? A: A multiple sequence alignment of a functionally diverse protein family and a representative PDB structure of one of the family members.
Q: Where can I get a multiple sequence alignment for my protein family? A: Use the Mustguseal server. Mustguseal can automatically construct large multiple alignments of protein families by collecting all available information about their structures and sequences in public databases. Multiple alignments of thousands of protein sequences and structures can be constructed using that public web-server.
Q: What do I get from pocketZebra? A: You get a list of binding sites in your protein ranked by functional significance and a list of positions in the structure that are important for selective binding of substrates/inhibitors/effectors. Both on-site and off-line versions of the output are provided.
Q: How can I cite pocketZebra? A: Please cite Suplatov D., Kirilin E., Arbatsky M., Takhaveev V., Švedas V. (2014) pocketZebra: a web-server for automated selection and classification of subfamily-specific binding sites by bioinformatic analysis of diverse protein families. Nucl. Acids Res. 42(W1): W344-W349. DOI:10.1093/nar/gku448
Q: What is a JobID? A: pocketZebra algorithm will be applied to your input data and your results will be placed in a public directory protected by a unique 14-symbol access code (JobID). JobID can be shared with a colleague and used to access your results at any time. You can always delete your results from the public directory by pressing the "Delete job" button at the top of the results page or by sending us an e-mail request containing the JobID.
Q: I have problems starting the interactive 3D structure viewer at the results page A: pocketZebra on-line interactivity at viewing the results is currently disabled.
