Zebra2
Advanced and easy-to-use web-server for bioinformatic analysis of subfamily-specific
and conserved positions in diverse protein superfamilies
This web-server is free and open to all users with no login requirement

... to identify and prioritize subfamily-specific and conserved positions in a functionally diverse superfamily
and to select hot-spots for rational design of the query protein
Version 2.0 since November 20th, 2019
THIS WEB-SITE/WEB-SERVER/SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS OR HARDWARE OWNERS OR WEB-SITE/WEB-SERVER/SOFTWARE MAINTEINERS/ADMINISTRATORS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE WEB-SITE/WEB-SERVER/SOFTWARE OR THE USE OR OTHER DEALINGS IN THE WEB-SITE/WEB-SERVER/SOFTWARE. PLEASE NOTE THAT OUR WEBSITE COLLECTS STANDARD APACHE2 LOGS, INCLUDING DATES, TIMES, IP ADDRESSES, AND SPECIFIC WEB ADDRESSES ACCESSED. THIS DATA IS STORED FOR APPROXIMATELY ONE YEAR FOR SECURITY AND ANALYTICAL PURPOSES. YOUR PRIVACY IS IMPORTANT TO US, AND WE USE THIS INFORMATION SOLELY FOR WEBSITE IMPROVEMENT AND PROTECTION AGAINST POTENTIAL THREATS. BY USING OUR SITE, YOU CONSENT TO THIS DATA COLLECTION.
Publication: D. Suplatov, Y. Sharapova, E. Geraseva, V. Švedas (2020) Zebra2: advanced and easy-to-use web-server for bioinformatic analysis of subfamily-specific and conserved positions in diverse protein superfamilies. Nucleic Acids Res., 48 (W1), W65–W71. DOI: 10.1093/nar/gkaa276
Navigation:
- Prerequisites and compatibility
- Preparing the input data for Zebra
- Description of parameters
- Tutorial to operate the results
- Examples of Zebra and interpretation of the results
- The Zebra file sharing and security features
- Citing Zebra
Highlights:
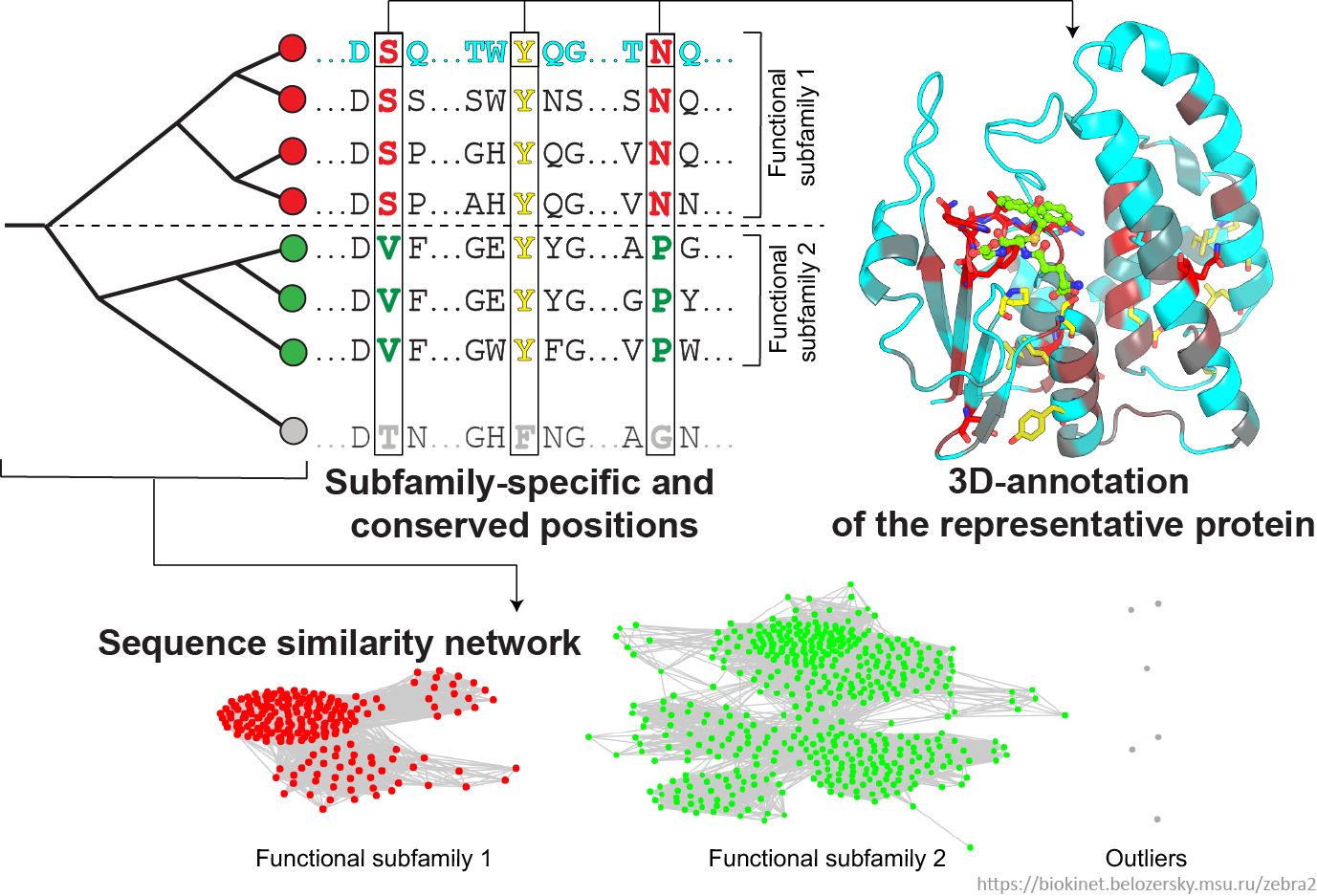
- The Zebra2 (i.e., Zebra v.2) is a highly automated web-tool to search for subfamily-specific and conserved positions (i.e., the determinants of functional diversity as well as the key catalytic and structural residues) in protein superfamilies;
- Information about both conserved and specific positions can help to understand how the enzyme performs its natural function, while the latter can also be selected as hotspots for directed evolution or rational design experiments in an attempt to improve the wild-type protein variant for a particular purpose;
- The bioinformatic analysis is supported by the integrated Mustguseal web-server to construct large structure-guided sequence alignments of functionally diverse families that can include thousands of proteins based on all available information in public databases;
- The Zebra2 results are automatically prioritized by the bioinformatic analysis and provided at four information levels to facilitate the knowledge-driven expert selection of the most promising positions on-line: as a sequence similarity network; interfaces to sequence-based and 3D-structure-based analysis of conservation and variability; and accompanied by the detailed annotation of proteins accumulated from the integrated databases with links to the external resources (i.e., PDB, Uniprot, BacDive, BRENDA);
- The integration of web-based bioinformatic tools Zebra2 and Mustguseal provides the first out-of-the-box open-access solution to identify subfamily-specific and conserved positions in protein superfamilies – to study function-associated patterns of local structure, to assist at selecting hot-spots for rational design and to prepare focused libraries for directed evolution.