Zebra3D Examples
Zebra3D can be used to identify the 3D-determinants of functional diversity in homologous proteins sharing a common fold of the superfamily, to assist protein engineering and help to annotate/study potential drug target sites/subsites that implement plasticity to facilitate accommodation of ligands. This page provides a brief overview of the Zebra3D+Mustguseal case-studies and further hosts download links to the complete data package with the respective input and output files.
Brief overview of the case-studies
Description and discussion of the ten case-studies illustrating diverse application of the Zebra3D+Mustguseal tool, including interpretation of the results, is provided in the Zebra3D publication. In brief, bioinformatic analysis of H1N1 Neuraminidase, Ornithine Decarboxylase from Trypanosoma brucei, Common ancestor of Haloalkane Dehalogenases and Renilla Luciferase, Polyester Hydrolase from Pseudomonas aestusnigri, Human Aldose Reductase, Guanine Deaminase, p38α MAP Kinase and HSP90, Malate Dehydrogenase from Sus scrofa, and Zinc metallo-beta-lactamase from Bacillus cereus and their corresponding superfamilies was carried out by the Zebra3D+Mustguseal. All input alignments for the discussed case-studies were created automatically by the Mustguseal web-server with the default settings starting from query PDB structures of the named enzymes. The analysis identified SSRs featuring dynamic loops and structural segments in the active site that were different among homologs and were previously established as key functionally important structural element in these enzymes involved in catalysis and/or substrate-recognition. The selected SSRs were previously used to study the 3D-determinants of catalytic activity and specific accommodation of ligands, helped to prepare focused libraries for directed evolution or assisted the design of chimeric enzymes with novel properties by exchange of equivalent regions between homologs, and characterized plasticity in binding sites to assist drug discovery efforts.
Quick example
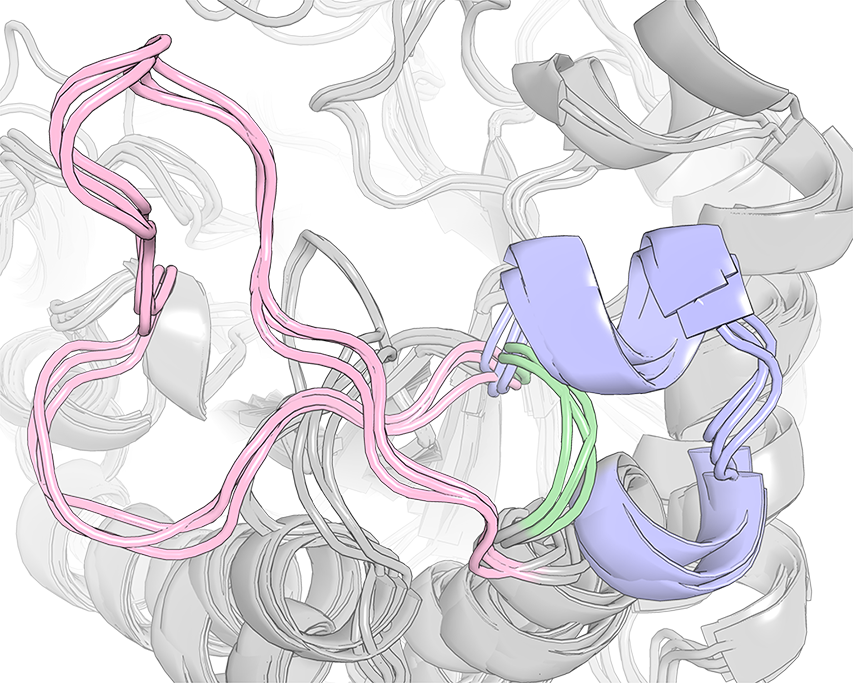
The case-study of Human Aldose Reductase and selected members of the Aldo-Keto Reductase superfamily is included with Zebra3D distribution package. See file 002_QUICK_EXAMPLE.txt from that package for instructions. As an example, the figure above shows an SSR #1 (i.e. ranked first out of 14) in 3D-alignment of selected members of the Aldo-Keto Reductase superfamily. This SSR #1 represented a region positioned at the top of the canonical (α/β)8-barrel structure that is significantly different between homologs featuring a diverse substrate specificity. Introduction of the corresponding flexible loop from Human Aldose Reductase (colored pink) instead of the corresponding fragment in structure of hyperthermostable Alcohol Dehydrogenase D from Pyrococcus furiosus (which was equivalent to those colored green), was one of the key steps in the creation of a chimera that implemented substrate specificity of the donor enzyme and inherited thermostability of the parent enzyme [10.1093/protein/gzs095].
Download all case-studies data
The input data (i.e. 3D-alignments created by the Mustguseal) and the results of Zebra3D analysis can be downloaded using the links below
| Input data only | download | [50 MB] | |
| Input data and Zebra3D results | download | [683 MB] |
The data is distributed as a 'tar.gz' archive pack. In Linux, use command tar xzf file.tar.gz to extract the data (both tar and gzip utilities usually come with the Linux distribution). In Windows, you can use a free tool 7-zip to unpack the archive.