Yosshi example
The Yosshi+Mustguseal integrated tool was used to study disulfide connectivity in subtilisin-like serine proteases, globins, lipases, carbonic anhydrases, xylanases, and members of the ribonuclease A superfamily. We conclude that introduction of S-S bonds identified by the bioinformatic analysis into the query protein structure can be successfully employed to improve properties of useful enzymes and design advanced proteins with controllable functions. The input and output data regarding these examples is provided below and briefly explained. A more detailed discussion is provided in the Yosshi publication.
- Example 1: Improving stability of subtilisin E from Bacillus subtilis
- Example 2: Design of Sperm Whale Myoglobin with a controllable function
- Download all case-studies as a single archive
Example 1: Improving stability of subtilisin E from Bacillus subtilis
NB: Full access to the input and output data regarding this example can be obtained by activating the “Demo mode” at the Yosshi submission page.
The aim. To introduce stabilizing disulfide bonds into the structure of subtilisin E from Bacillus subtilis
The strategy. Lets study the homologs of subtilisin - the stabilizing disulfide may already be present in structures of these proteins, i.e., it is absent from the selected subtilisin but can be naturally occurring in thermophilic homologs within the superfamily which share the common structural fold with our query.
Construction of the multiple alignment. The Mustguseal web-server with parameters set according to the "Scenario 3" was used to construct an alignment of a large representative set of proteins from the subtilisin-like serine proteases superfamily. The PDB code 1SCJ (chain A) corresponding to the structure of subtilisin E from Bacillus subtilis (i.e., the query/representative protein) was submitted in the Mode 1 as discussed here. As a result, an alignment of a non-redundant set of 8456 sequences and structures of proteins with high structural but low sequence similarity to the query protein was automatically constructed. The alignment can be downloaded using the link below.
- Download the alignment automatically created by the Mustguseal containing 8456 sequences and structures of proteins with high structural but low sequence similarity to the subtilisin E from Bacillus subtilis: [download]
- Download the PDB structure of subtilisin E from Bacillus subtilis: [download]
The analysis by Yosshi. The automatically prepared multiple alignment and the PDB structure of subtilisin E from Bacillus subtilis was further submitted to Yosshi for analysis with the default settings. Processing of the data took less than 5 minutes. The Yosshi results can be downloaded using the links below. The content of these files and guidelines to study them are provided on this page.
- Download the Yosshi annotation of the representative protein structure according to the bioinformatic and structural analysis of homologs: [download]
- Download the text version of the Yosshi annotation: [download]
- Download the Yosshi PyMol script pack: [download]
- Download the 3D-models of mutants of the representative protein with incorporated S-S bonds: [download]
Interpretation of the Yosshi results. The Yosshi results can be downloaded to your computer for a local analysis or studied on-line using the HTML5-based interactive analysis tools. The information presented at the on-line analysis page is equivalent to the PyMol PSE structural annotation file and the text summary of the annotation. See this page for a detailed discussion of the Yosshi output.
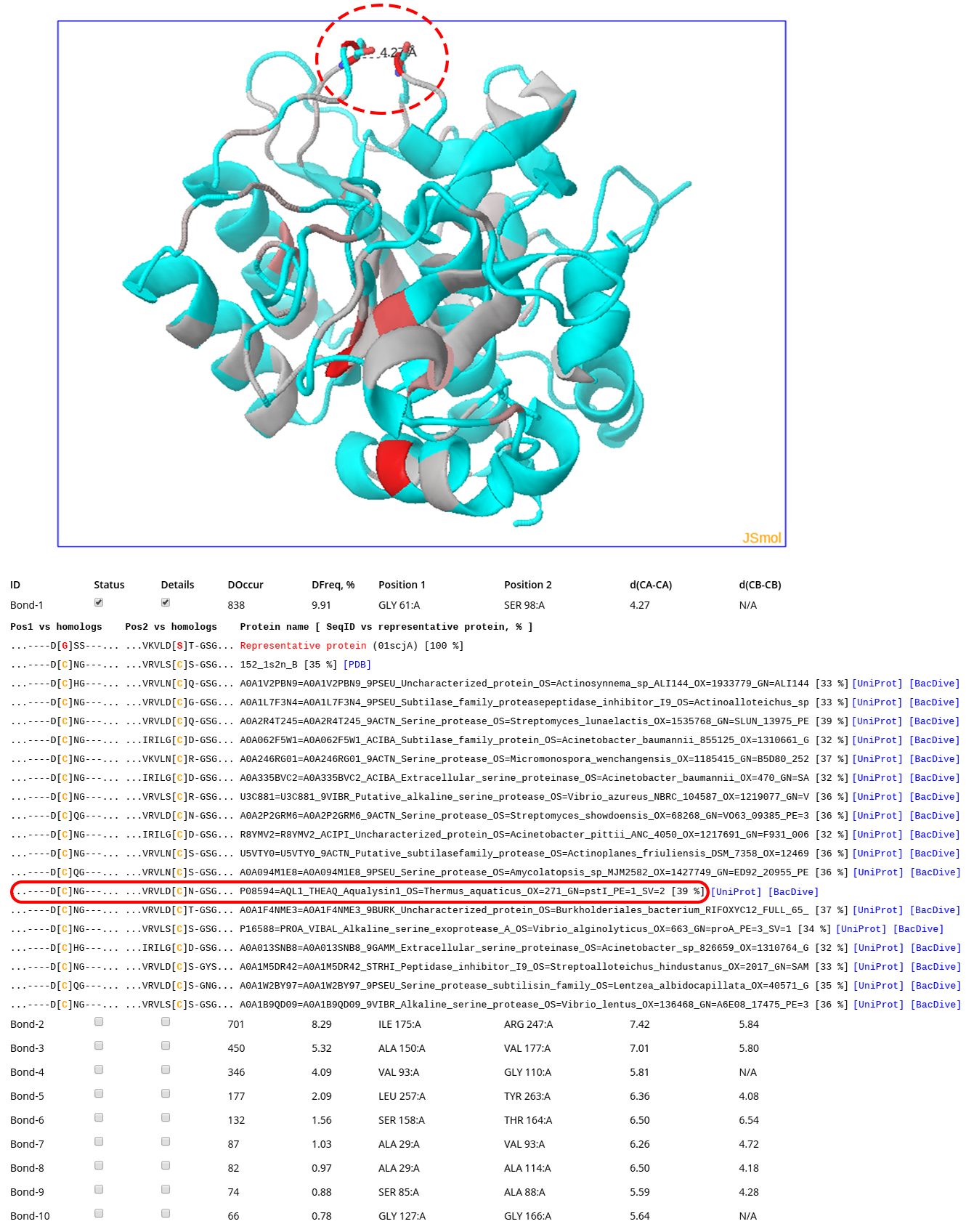
Click here to enlarge
The screenshot above was taken at the on-line analysis page for the subtilisin example. To generate that output the "Provide HTML links to PDB, UniProt, and BacDive" feature was turned ON (see this page for details). The structure of subtilisin E from Bacillus subtilis (PDB 1SCJ) is shown in the 3D viewer. The backbone of each pair of positions in the structure of the query protein in the 3D-viewer is gradient painted from grey to red according to the DOccur (i.e., "Disulfide Occurrence") - the number of times both positions are occupied by Cysteines in sequences and structures of homologs in the multiple alignment (i.e., the expected occurrence of the corresponding crosslink in protein families). In total, 56 pairs of positions in the query protein structure were selected as capable of a disulfide bond formation upon mutation to Cysteines, and ranked based on the expected abundance of the corresponding crosslink in homologs. The #1 prediction corresponded to a pair of residues Gly61 and Ser98 which are occupied by Cysteines in 838 proteins (shown as sticks and indicated by a dashed oval). The corresponding sub-sequences of some of these homologs are shown (Cysteines are colored in yellow). Cysteines in both positions can be observed in proteins from thermophilic organisms, e.g. Aqualysin-1 from Thermus aquaticus (indicated by a red oval). The provided link to the BacDive entry for Thermus aquaticus includes an annotated thermophilic temperature range for this organism (e.g., growth at 70 C) what is significantly higher compared to that of Bacillus subtilis (i.e., growth at 30 C). Introduction of this crosslink which naturally occurs in thermophilic organisms into the structure of subtilisin E from Bacillus subtilis by a double mutation Gly61Cys/Ser98Cys was previously reported to significantly enhance thermostability without changing catalytic efficiency of the enzyme (https://doi.org/10.1093/protein/9.9.789). Another 55 pairs of positions selected by the bioinformatic analysis in the query protein were found to be substituted by Cysteines in up to 701 homologs with different properties and originating from different organisms, and thus provide a list of promising candidate hot-spots to further engineer subtilisin and its evolutionary relatives.
Example 2: Design of Sperm Whale Myoglobin with a controllable function
The Yosshi+Mustguseal integrated on-line tool was used to study the Globins superfamily. The PDB structure 1JP6 (chain A) of Sperm Whale Myoglobin was submitted to the Mustguseal web-server to automatically construct an alignment of a non-redundant set of 5554 proteins with high structural but low sequence similarity to the query. A pair of positions Val21 and Val66 in the Myoglobin structure was ranked #1 out of 21 candidates and was substituted by Cysteines in 78 protein sequences annotated as Cytoglobines. Analysis of the literature showed that the S-S bond between equivalent residues in Cytoglobines regulates protein reactivity by reshaping the internal cavities and thus modulating the mechanism of CO escape, although no crystallographic structure with this disulfide in the oxidized (i.e., bonded) form is currently available. Introduction of this naturally occurring crosslink into the structure of Sperm Whale Myoglobin by a double mutation Val21Cys/Val66Cys was previously reported to implement a regulatory mechanism to fine-tune the catalytic reactivity of the protein (https://doi.org/10.1039/c8cc01646a).
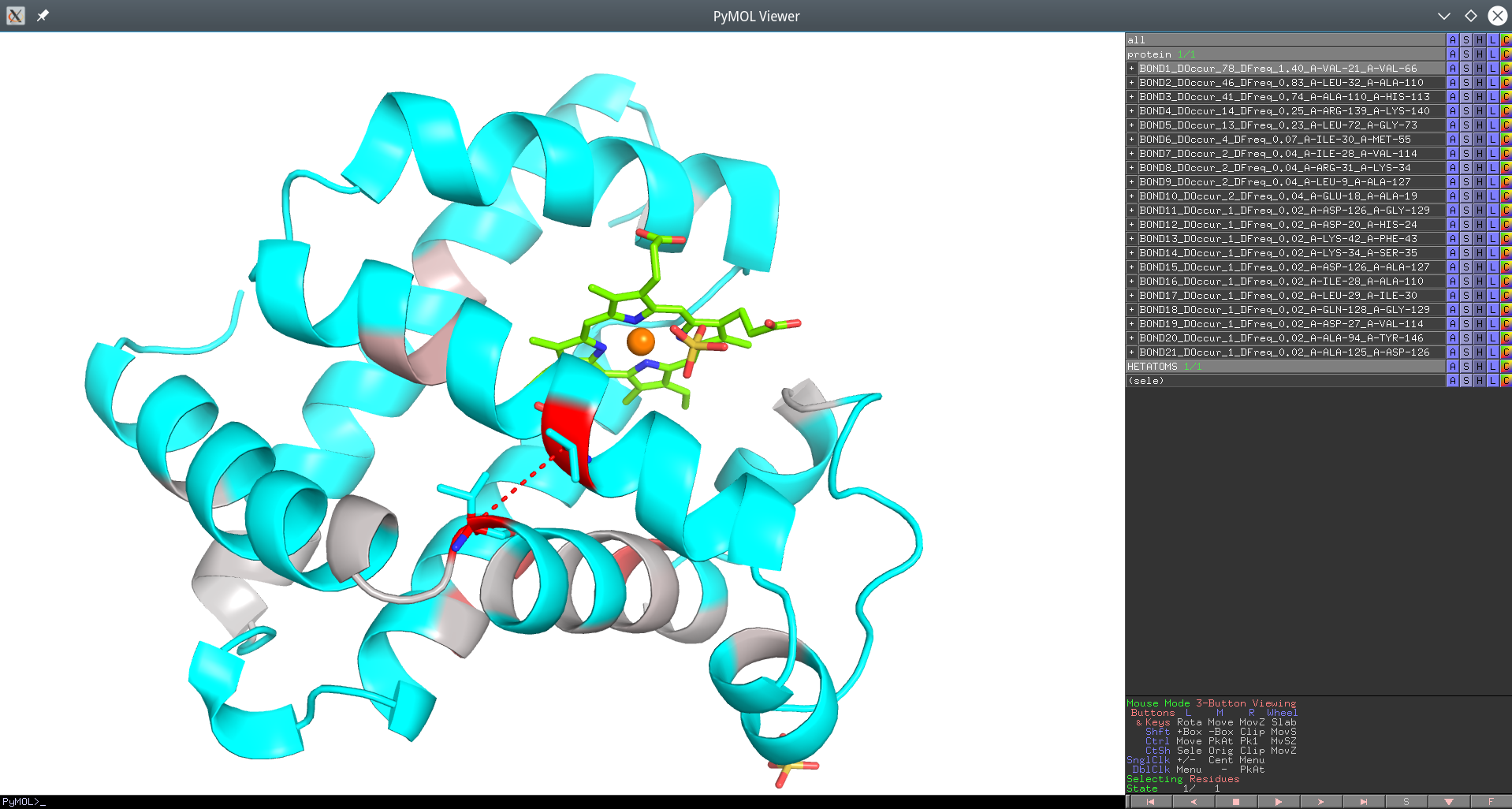
Click here to enlagre
Yosshi homology-based annotation of the Sperm Whale Myoglobin according to presence of disulfides in the Globins superfamily provided as a PyMol PSE session file. The #1 pair of positions Val21 and Val66 which is the focus of this example is connected by a red dotted line.
The automatically created structure/sequence alignment of the Globins superfamily and the Yosshi analysis files regarding this example are provided below:
- Download the alignment automatically created by the Mustguseal containing 5554 sequences and structures of proteins with high structural but low sequence similarity to the Sperm Whale Myoglobin: [download]
- Download the PDB structure of Sperm Whale Myoglobin: [download]
- Download the Yosshi annotation of the representative protein structure according to the bioinformatic and structural analysis of homologs: [download]
- Download the text version of the Yosshi annotation: [download]
- Download the Yosshi PyMol script pack: [download]
- Download the 3D-models of mutants of the representative protein with incorporated S-S bonds: [download]
Download all case-studies as a single archive
In addition to the bioinformatic analysis of subtilisin and myoglobin and their corresponding superfamilies discussed above, more case-studies of different lipases, carbonic anhydrases, xylanases, and members of the ribonuclease A superfamily were performed to reproduce the previously reported disulfide engineering experiments. The input and output data regarding all case-studies can be downloaded as a single files: [download] (33 MB). A detailed discussion is provided in the Yosshi publication.