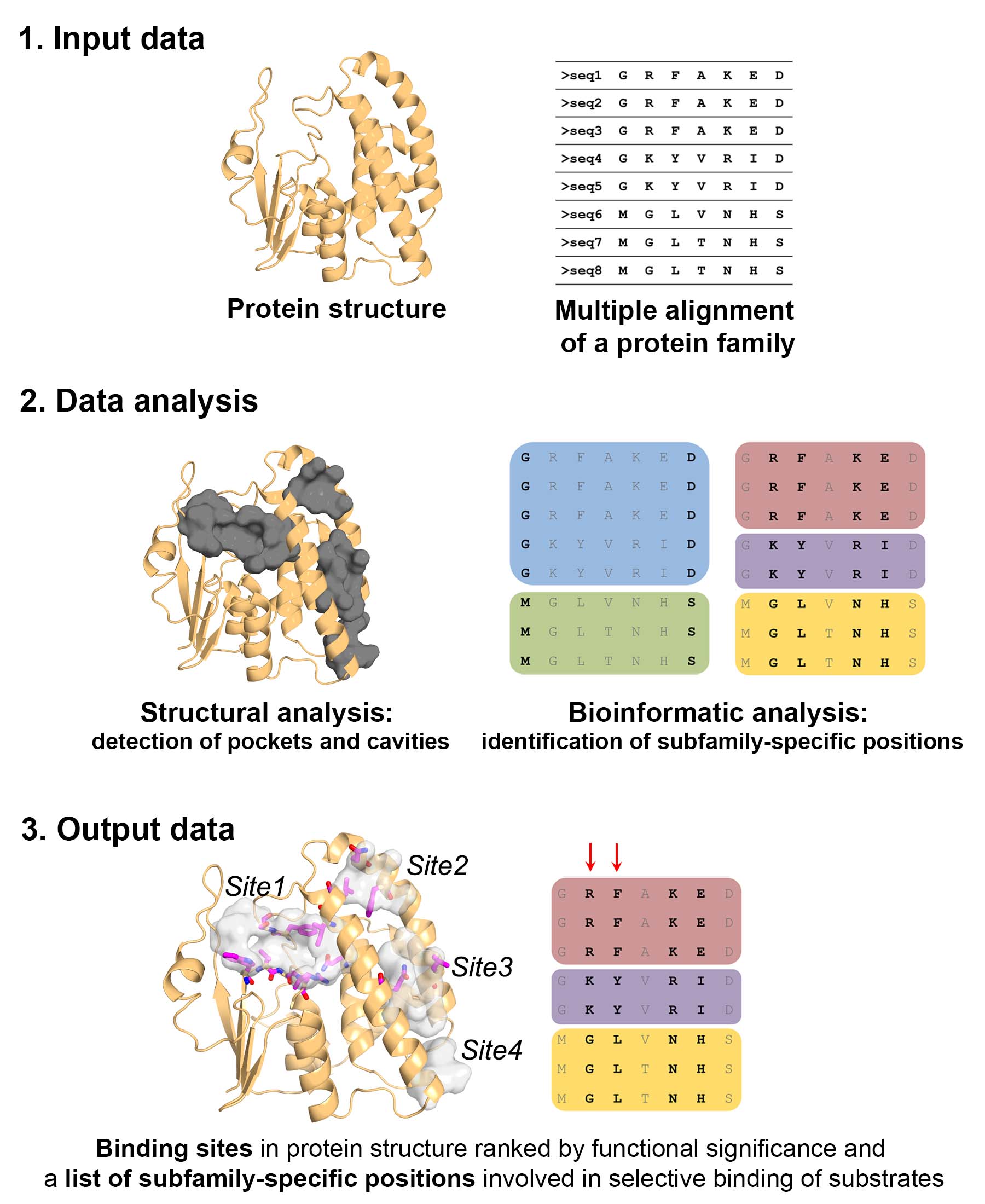
The pocketZebra Algorithm
pocketZebra algorithm includes three basic steps:
- First, pocketZebra performs the bioinformatic analysis of distant protein families to identify subfamily-specific positions – conserved within functional subfamilies, but different between subfamilies. For the bioinformatic analysis pocketZebra implements a recently developed algorithm Zebra (Suplatov et al., 2020; learn more about Zebra).
- The second step is the structure-based detection of cavities in proteins – potential functionally important sites involved in intermolecular binding. This step by default is performed with the Fpocket algorithm (Schmidtke et al., 2010). Alternatively any locally installed algorithm can be implemented by user and the resulting pocket definition then uploaded to the server.
- Finally, the subfamily-specific positions are mapped to the identified structural cavities and statistical analysis is performed to select the most significant subfamily-specific binding sites and rank them by the presence of the subfamily-specific positions to suggest functional importance.
The algorithm of pocketZebra is described in more detail in Suplatov D., Kirilin E., Arbatsky M., Takhaveev V., Švedas V. (2014) pocketZebra: a web-server for automated selection and classification of subfamily-specific binding sites by bioinformatic analysis of diverse protein families. Nucl. Acids Res. 42(W1): W344-W349. DOI:dx.doi.org