Yosshi output
The output of Yosshi is a detailed homology-based annotation of the user-submitted query protein structure describing the occurrence of disulfides within a common structural fold of a superfamily. This annotation can be used to study disulfide connectivity in homologs with different properties, as well as to identify disulfide bridges present in homologs but not in the query protein that can be introduced to design its stability and functional properties. The most abundant (i.e., conserved) disulfides are ranked first to facilitate their further analysis in particular proteins, as presence of an S-S bond in a larger group of proteins within a superfamily suggests its direct role in a function or property common among these homologs. The results of the Yosshi web-server can be downloaded to your computer for local analysis or studied on-line using the built-in interactive analysis tools. Interactivity is implemented in HTML5 and therefore neither plugins nor Java are required.
- Residue numbering in the Yosshi output
- General description
- Download section
- The Yosshi annotation
- On-line analysis section
Residue numbering in the Yosshi output
Please note, that the residue numbering in the Yosshi output may differ from the numbering in the PDB file that you have originally submitted. E.g., amino acid residues with identical IDs within the same chain will be identified and renumbered automatically. The complete list of changes to your query PDB will be printed to the log file. The most important changes will be colored in red. Press the "Log" button to view the on-line log page. E.g.:
Warning: Amino acid residues with identical residue IDs have been identifies and will be renumbered as described below
Warning: Residues A-TYR-169 and A-PRO-169 have the same residue ID
Warning: Offseting all residue IDs starting from 169 in chain A by 1
Warning: Residues A-HIS-413 and A-GLN-413 have the same residue ID
Warning: Offseting all residue IDs starting from 413 in chain A by 2
Warning: Residues A-PRO-414 and A-HIS-414 have the same residue ID
Warning: Offseting all residue IDs starting from 414 in chain A by 3
Info: Correcting the PDB file...
The corrected PDB file of your query can be downloaded at the "Results" page, see section "The Input data after preprocessing".
General description
The first output is a list of pairs of positions in the structure of the query protein that can form a disulfide bond assuming both residues are mutated to cysteines, or that already are occupied by cysteines which can form a crosslink. The second output is a list of homologs of the query protein, which contain cysteines in equivalent positions for each pair of such residues. For each such homolog and its source organism the HTML links to respective pages in PDB, UniProt, and BacDive can be automatically created to facilitate further expert analysis (please see this page for more information about the "Provide HTML links to PDB, UniProt, and BacDive" feature and its current limitations). The selected pairs are ranked in a descending order of the “Disulfide Occurrence” (DOccur) what is a positive integer equal to the number of times they are both occupied by cysteines in sequences of homologs (i.e., the expected occurrence of corresponding crosslink in protein families). In addition, the “Disulfide Frequency” (DFreq) is provided for each pair of positions that takes a value from the range 0-100% calculated as DOccur divided by the total number of proteins in the multiple alignment. The interpretation of the DOccur metric is equivalent to that of the popular subfamily-specific conservation. A higher value of the DOccur indicates that a corresponding disulfide is conserved in a larger group of proteins within a superfamily suggesting its direct role in a function or property common among these homologs (e.g., in structural stability or a regulatory mechanism, as discussed here). Therefore, the purpose of the Yosshi ranking is to show the most conserved disulfides first to facilitate their further analysis in particular proteins. The Yosshi results are primarily web-based and viewable on-site, but can also be downloaded to a local computer.
Download section

The Download section provides download links to the primary output (the query protein structure annotated according to the selected pairs of positions capable of a disulfide bond formation upon mutation to Cysteines or that already are occupied by Cysteines which can form a crosslink, ranked based on the expected abundance of the corresponding S-S bond in homologs), supplementary output (a text file listing the selected pairs of positions, the PyMol script pack to reproduce the primary output on your local computer), and the user input data after preprocessing by the Yosshi server (the multiple sequence alignment and the representative protein structure).
- The Yosshi annotation file is a PyMol 'PSE' session file which contains the representative (query) protein structure annotated according to the selected pairs of positions capable of a disulfide bond formation upon mutation to Cysteines (or that already are occupied by Cysteines which can form a crosslink), ranked based on the expected abundance of the corresponding S-S bond in homologs. The details of the Yosshi annotation are discussed below. To open this 'PSE' session file you will need PyMol v. 1.7.3.0 or higher. If you have an older version of PyMol you may need to recompile the PSE session by manually executing the PyMol script, which was created by the Yosshi web-server (see "The Yosshi PyMol script pack" below).
- The text version of the Yosshi annotation is a plain text file listing the selected pairs of residues in the representative protein structure and details about names and sequences of homologs which were discovered to contain Cysteines in the equivalent positions. This text file also contains the information about the total number of proteins in the submitted alignment. The content of the file is optimized for automatic processing by third-party scripts (i.e., header-based text parsing). An example of the text summary file is provided below.
- The Yosshi PyMol script pack contains all information required to recompile the Yosshi annotation 'PSE' file. The pack is provided in the unlikely event of incompatibility of the 'PSE' file compiled by the Yosshi web-server with the PyMol version installed on the user`s local computer. The files are packed in 'tar.gz' archive. To extract files from a 'tar.gz' archive use the command
tar xzf yosshi_[TaskID].tar.gzin Linux and in Widows use a free 7-zip tool. To recompile the 'PSE' session run the commandpymol -c yosshi_[TaskID]_pymol.pyin Linux or by using"File" → "Run Script"interface menu in Windows. Would your PyMol version fail to compile the Yosshi annotation from the provided script please contact support.
Yosshi annotation
In general, for each pair of positions selected by Yosshi in the query (representative) protein structure (i.e., occupied by a native S-S bonds, or a promising site for S-S bond formation assuming both positions are mutated to cysteines) the following information is provided:
- the value of “Disulfide Occurrence” (DOccur), i.e., the expected occurrence of the corresponding crosslink in protein families;
- the “Disulfide Frequency” (DFreq) which takes a value from the range 0-100% calculated as DOccur divided by the total number of proteins in the multiple alignment;
- a list of homologs that seem to contain a disulfide bond in equivalent positions as this information can help to interpret their roles in protein families;
- the list of high-resolution PDB structures (i.e., not necessarily homologs to the query) that were used to create the 3D-motif library and contain two covalently connected cysteines that can fit into the selected positions in the query structure with respect to the chosen statistical model, i.e., the PDB records that contain the 3D-motifs that matched with the pair of candidate positions selected by Yosshi in the query protein structure.
The details are provided below.
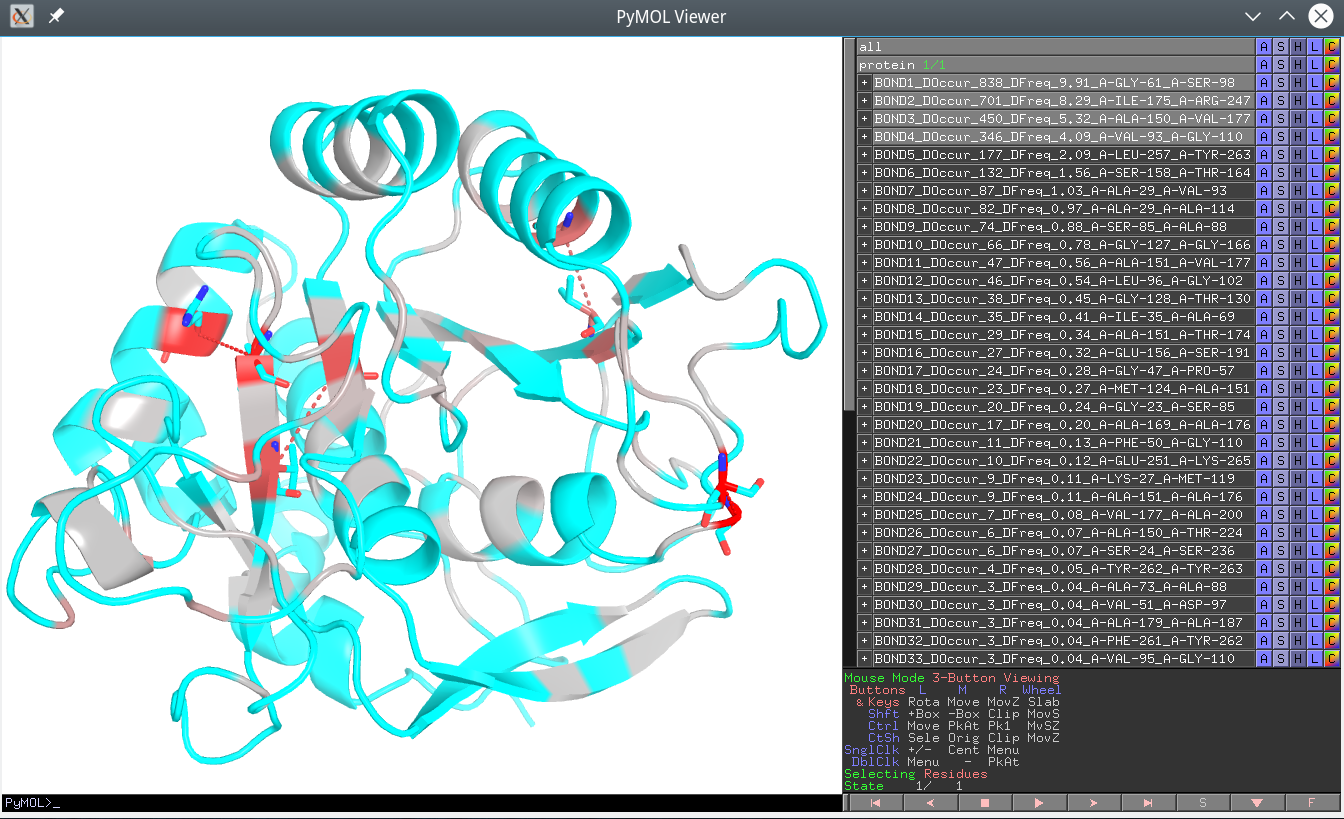
Click here to enlagre
The Yosshi output is a homology-based annotation of native S-S bonds in the query (representative) protein structure as well as the disulfide bridges present in homologs, but not in the query protein. These pairs are ranked in a descending order of the “Disulfide Occurrence” (DOccur), i.e., the expected occurrence of the corresponding crosslink in protein families. A higher value of the DOccur indicates that a corresponding disulfide is conserved in a larger group of proteins within a superfamily suggesting its direct role in a function or property common among these homologs (e.g., in structural stability or a regulatory mechanism, as discussed below). Therefore, the purpose of the Yosshi ranking is to show the most conserved disulfides first to facilitate their further analysis in particular proteins. In addition, the “Disulfide Frequency” (DFreq) is provided for each pair of positions that takes a value from the range 0-100% calculated as DOccur divided by the total number of proteins in the multiple alignment.
In the Yosshi PyMol annotation file (i.e., the off-line version of the output) each selected pair of positions in the representative protein structure will be shown as sticks, connected by a dashed line, and provided as a separate object of the name
BOND[rank]_DOccur_[integer]_DFreq_[float]_[Position-1]_[Position-2]. Click on the name of the object in the Pymol panel to enable/disable the corresponding bond in the 3D-viewer.
The Yosshi PyMol annotation file should be studied together with the text version of the Yosshi annotation file, which contains a list of the selected pairs of residues and details about names and sequences of homologs which were discovered to contain cysteines in the equivalent positions. This text file also contains the information about the total number of proteins in the submitted alignment. If the "Provide HTML links to PDB, UniProt, and BacDive" was turned on, then the corresponding links will be provided for each protein at the end of each line. The content of the file is optimized for automatic processing by third-party scripts (i.e., header-based text parsing). See an example below (a cropped version of a real file):
# YOSSHI OUTPUT SUMMARY FOR TASK yosshi_zy1qok75sb73ra #
# The first output is a list of pairs of positions in the structure of the query protein that can form a disulfide bond assuming both residues # are mutated to Cysteines, or that already are occupied by Cysteines which can form a crosslink. The second output is a list of homologs of
# the query protein, which contain Cysteines in equivalent positions for each pair of such residues. The selected pairs are ranked in a
# descending order of the "Disulfide Occurrence" (DOccur). The DOccur for a pair of positions in the structure of a query protein is a positive
# integer equal to the number of times they are occupied by Cysteines in sequences of homologs (i.e., the expected occurrence of corresponding
# crosslink in protein families). In addition, the "Disulfide Frequency" (DFreq) is provided for each pair of positions that takes a value from
# the range 0-100% calculated as DOccur divided by the total number of proteins in the multiple alignment. # # For each BOND the following information is provided:
# * SUMMARY - description of the selected positions POS1 and POS2 in the query/representative protein structure;
# DOccur: the "Disulfide Occurrence" (see the description above);
# DFreq: the "Disulfide Frequency" (in %, see the description above);
# PDB1 and PDB2: numbering of the two positions according to the query/representative protein structure;
# ALN1 and ALN2: numbering of the two positions according to their column IDs in the multiple sequence alignment (starting from 1);
# D(CA-CA) and D(CB-CB): distances between the respective atoms of residues POS1 and POS2 in the query/representative protein structure;
# * DETAILS - description of each PROTEIN from the alignment which contains Cysteines in the equivalent positions;
# ALN1 and ALN2: sequence fragments of the homologous PROTEIN aligned to POS1 and POS2 of the representative protein structure;
# The residues actually aligned to POS1 and POS2 are shown in square brackets; # PROTEIN: The complete protein name as in the alignment file truncated to the first 99 characters. # Sequence identity of the PROTEIN with the query/representative protein is given in square brackets.
# Each BOND line ends with an END keyword # TOTAL_PROT 8456 (the number of proteins in the multiple alignment which was submitted for the analysis)
# BOND1 SUMMARY DOccur: 838 DFreq: 9.91 % POS1: [PDB: A GLY 61 ] [ALN: 282 ] POS2: [PDB A SER 98 ] [ALN: 346 ] D(CA-CA): 4.27 D(CB-CB): N/A END BOND1 DETAILS ALN1: ...----D[G]SS---... ALN2: ...VKVLD[S]T-GSG... PROTEIN: Representative protein (01scjA) [100 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...VRVLS[C]S-GSG... PROTEIN: 152_1s2n_B [35 %] END
BOND1 DETAILS ALN1: ...----D[C]HG---... ALN2: ...VRVLN[C]Q-GSG... PROTEIN: A0A1V2PBN9=A0A1V2PBN9_9PSEU_Uncharacterized_protein_OS=Actinosynnema_sp_ALI144_OX=1933779_GN=ALI144 [33 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...VRVLD[C]G-GSG... PROTEIN: A0A1L7F3N4=A0A1L7F3N4_9PSEU_Subtilase_family_proteasepeptidase_inhibitor_I9_OS=Actinoalloteichus_sp [33 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...VRVLD[C]Q-GSG... PROTEIN: A0A2R4T245=A0A2R4T245_9ACTN_Serine_protease_OS=Streptomyces_lunaelactis_OX=1535768_GN=SLUN_13975_PE [39 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...IRILG[C]D-GSG... PROTEIN: A0A062F5W1=A0A062F5W1_ACIBA_Subtilase_family_protein_OS=Acinetobacter_baumannii_855125_OX=1310661_G [32 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...VKVLN[C]R-GSG... PROTEIN: A0A246RG01=A0A246RG01_9ACTN_Serine_protease_OS=Micromonospora_wenchangensis_OX=1185415_GN=B5D80_252 [37 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...IRILG[C]D-GSG... PROTEIN: A0A335BVC2=A0A335BVC2_ACIBA_Extracellular_serine_proteinase_OS=Acinetobacter_baumannii_OX=470_GN=SA [32 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...VRVLS[C]R-GSG... PROTEIN: U3C881=U3C881_9VIBR_Putative_alkaline_serine_protease_OS=Vibrio_azureus_NBRC_104587_OX=1219077_GN=V [36 %] END
BOND1 DETAILS ALN1: ...----D[C]QG---... ALN2: ...VRVLD[C]N-GSG... PROTEIN: A0A2P2GRM6=A0A2P2GRM6_9ACTN_Serine_protease_OS=Streptomyces_showdoensis_OX=68268_GN=VO63_09385_PE=3 [36 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...IRILG[C]D-GSG... PROTEIN: R8YMV2=R8YMV2_ACIPI_Uncharacterized_protein_OS=Acinetobacter_pittii_ANC_4050_OX=1217691_GN=F931_006 [32 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...VRVLN[C]S-GSG... PROTEIN: U5VTY0=U5VTY0_9ACTN_Putative_subtilasefamily_protease_OS=Actinoplanes_friuliensis_DSM_7358_OX=12469 [36 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...VRVLS[C]S-GSG... PROTEIN: A0A2M7LPT2=A0A2M7LPT2_9GAMM_Alkaline_serine_protease_OS=Shewanella_sp_CG_4_10_14_3_um_filter_42_91_ [35 %] END
BOND1 DETAILS ALN1: ...----D[C]NG---... ALN2: ...VRVLN[C]S-GSG... PROTEIN: A0A1C6RXP9=A0A1C6RXP9_9ACTN_Peptidase_inhibitor_I9_OS=Micromonospora_inyonensis_OX=47866_GN=GA00746 [35 %] END
... ... ...
BOND2 SUMMARY DOccur: 701 DFreq: 8.29 % POS1: [PDB: A ILE 175 ] [ALN: 556 ] POS2: [PDB A ARG 247 ] [ALN: 733 ] D(CA-CA): 7.42 D(CB-CB): 5.84 END
BOND2 DETAILS ALN1: ...---ST[I]AVGAV... ALN2: ...---QV[R]DRLES... PROTEIN: Representative protein (01scjA) [100 %] END
BOND2 DETAILS ALN1: ...---NV[C]TIAAS... ALN2: ...----A[C]ARIVQ... PROTEIN: 171_5z6o_A [32 %] END
BOND2 DETAILS ALN1: ...---MA[C]TVGAT... ALN2: ...----L[C]NLIAS... PROTEIN: A0A2K0WUH3=A0A2K0WUH3_GIBNY_Uncharacterized_protein_OS=Gibberella_nygamai_OX=42673_GN=FNYG_00989_PE [31 %] END
BOND2 DETAILS ALN1: ...---SI[C]TVAAS... ALN2: ...----L[C]DTIKN... PROTEIN: E4V2V9=SUB7_ARTGP_Subtilisinlike_protease_7_OS=Arthroderma_gypseum_strain_ATCC_MYA4604__CBS_118893_ [32 %] END
BOND2 DETAILS ALN1: ...---NV[C]TIAAS... ALN2: ...----A[C]ARIVE... PROTEIN: Q6PLK0=Q6PLK0_PENCH_Alkaline_serine_protease_OS=Penicillium_chrysogenum_OX=5076_PE=3_SV=1 [31 %] END BOND2 DETAILS ALN1: ...---TV[C]TVGAS... ALN2: ...----V[C]DRMKE... PROTEIN: A0A2B7Y6K0=A0A2B7Y6K0_9EURO_Uncharacterized_protein_OS=Helicocarpus_griseus_UAMH5409_OX=1447875_GN= [34 %] END
BOND2 DETAILS ALN1: ...---SI[C]TVAAS... ALN2: ...----L[C]DTIKQ... PROTEIN: Q64K36=SUB7_ARTBE_Subtilisinlike_protease_7_OS=Arthroderma_benhamiae_OX=63400_GN=SUB7_PE=1_SV=1 [33 %] END
BOND2 DETAILS ALN1: ...---GV[C]AVGAS... ALN2: ...----L[C]ERIKE... PROTEIN: C5PFR5=SU11B_COCP7_Subtilisinlike_protease_CPC735_047380_OS=Coccidioides_posadasii_strain_C735_OX=2 [33 %] END
BOND2 DETAILS ALN1: ...---SV[C]TIAAS... ALN2: ...----A[C]LRLKQ... PROTEIN: C5G1D1=SUB5_ARTOC_Subtilisinlike_protease_5_OS=Arthroderma_otae_strain_ATCC_MYA4605__CBS_113480_OX= [33 %] END
BOND2 DETAILS ALN1: ...---KV[C]AVGAS... ALN2: ...----L[C]DRIKQ... PROTEIN: C4JSL8=C4JSL8_UNCRE_Uncharacterized_protein_OS=Uncinocarpus_reesii_strain_UAMH_1704_OX=336963_GN=UR [33 %] END
BOND2 DETAILS ALN1: ...---KV[C]TVSAT... ALN2: ...----L[C]DRLKQ... PROTEIN: A0A0J7AZ74=A0A0J7AZ74_COCIT_Subtilasetype_proteinase_psp3_OS=Coccidioides_immitis_RMSCC_2394_OX=404 [30 %] END
BOND2 DETAILS ALN1: ...---GV[C]AIAAS... ALN2: ...----V[C]TRIKE... PROTEIN: J3KC10=J3KC10_COCIM_Subtilisinlike_protease_OS=Coccidioides_immitis_strain_RS_OX=246410_GN=CIMG_037 [29 %] END
BOND2 DETAILS ALN1: ...---GV[C]AIAAS... ALN2: ...----V[C]NRIKE... PROTEIN: E9D0A9=E9D0A9_COCPS_Alkaline_protease_OS=Coccidioides_posadasii_strain_RMSCC_757__Silveira_OX=44322 [29 %] END
... ... ...
BOND3 SUMMARY DOccur: 450 DFreq: 5.32 % POS1: [PDB: A ALA 150 ] [ALN: 474 ] POS2: [PDB A VAL 177 ] [ALN: 558 ] D(CA-CA): 7.01 D(CB-CB): 5.80 END
BOND3 DETAILS ALN1: ...G-IVV[A]AAAGN... ALN2: ...-STIA[V]GAVNS... PROTEIN: Representative protein (01scjA) [100 %] END
BOND3 DETAILS ALN1: ...G-VTI[C]AASGD... ALN2: ...-YVLA[C]GGTTL... PROTEIN: A0A0P0KB00=A0A0P0KB00_9BURK_Serine_protease_kumamolysin_OS=Burkholderia_plantarii_OX=41899_GN=bpln_ [18 %] END
BOND3 DETAILS ALN1: ...G-VTV[C]VASGD... ALN2: ...-YVLA[C]GGTNL... PROTEIN: A0A221ABI4=A0A221ABI4_9BURK_Pseudomonalisin_OS=Burkholderia_sp_AD24_OX=1528693_GN=pcp_1_PE=4_SV=1 [19 %] END
BOND3 DETAILS ALN1: ...G-ITV[C]VASGD... ALN2: ...-YVLA[C]GGTRL... PROTEIN: A0A0H3I768=A0A0H3I768_PECSS_Peptidase_S53_propeptide_OS=Pectobacterium_sp_strain_SCC3193_OX=1905730 [16 %] END
BOND3 DETAILS ALN1: ...G-ITV[C]CAAGD... ALN2: ...-YALA[C]GGTQL... PROTEIN: A0A2V5VDV0=A0A2V5VDV0_9BACT_Peptidase_S53_OS=Verrucomicrobia_bacterium_OX=2026799_GN=DME69_05485_PE [17 %] END
BOND3 DETAILS ALN1: ...G-ITV[C]AASGD... ALN2: ...-YVLG[C]GGTQL... PROTEIN: B9BGB8=B9BGB8_9BURK_Serine_protease_subtilase_family_OS=Burkholderia_multivorans_CGD1_OX=513051_GN= [17 %] END
BOND3 DETAILS ALN1: ...G-VTV[C]VASGD... ALN2: ...-FALA[C]GGTNL... PROTEIN: A0A023XTZ1=A0A023XTZ1_BRAJP_Uncharacterized_protein_OS=Bradyrhizobium_japonicum_SEMIA_5079_OX=47628 [19 %] END
BOND3 DETAILS ALN1: ...G-VTV[C]AASGD... ALN2: ...-YALG[C]GGTSL... PROTEIN: A0A158BNR8=A0A158BNR8_9BURK_Peptidase_S53_propeptide_OS=Caballeronia_glebae_OX=1777143_GN=AWB82_043 [17 %] END
BOND3 DETAILS ALN1: ...G-VTV[C]AASGD... ALN2: ...-YVLG[C]GGTSL... PROTEIN: A0A157Z216=A0A157Z216_9BURK_Peptidase_S53_propeptide_OS=Caballeronia_glebae_OX=1777143_GN=AWB82_001 [17 %] END
BOND3 DETAILS ALN1: ...G-VTV[C]AASGD... ALN2: ...-YALA[C]GGTSV... PROTEIN: A0A1I5R9V1=A0A1I5R9V1_9RALS_Kumamolisin_OS=Ralstonia_sp_NFACC01_OX=1566294_GN=SAMN03159417_02427_PE [18 %] END
BOND3 DETAILS ALN1: ...G-VTV[C]VASGD... ALN2: ...-YVLA[C]GGTNL... PROTEIN: A0A1H1HLP5=A0A1H1HLP5_9BURK_Kumamolisin_Serine_peptidase_MEROPS_family_S53_OS=Paraburkholderia_fung [20 %] END
BOND3 DETAILS ALN1: ...G-VTI[C]AASGD... ALN2: ...-YVLA[C]GGTHL... PROTEIN: A0A1I9YLY5=A0A1I9YLY5_9BURK_Peptidase_S53_OS=Paraburkholderia_sprentiae_WSM5005_OX=754502_GN=BJG93_ [19 %] END
... ... ...
BOND56 SUMMARY DOccur: 1 DFreq: 0.01 % POS1: [PDB: A ILE 115 ] [ALN: 386 ] POS2: [PDB A ALA 142 ] [ALN: 465 ] D(CA-CA): 5.96 D(CB-CB): 5.33 END
BOND56 DETAILS ALN1: ...GIEWA[I]SNN--... ALN2: ...TVVDK[A]VSSG-... PROTEIN: Representative protein (01scjA) [100 %] END
BOND56 DETAILS ALN1: ...AIDLA[C]DAG--... ALN2: ...RAVRR[C]REQG-... PROTEIN: A0A1H7JF26=A0A1H7JF26_9ACTN_Cyanobactin_maturation_protease_PatAPatG_family_OS=Nonomuraea_pusilla_O [28 %] END
EOF
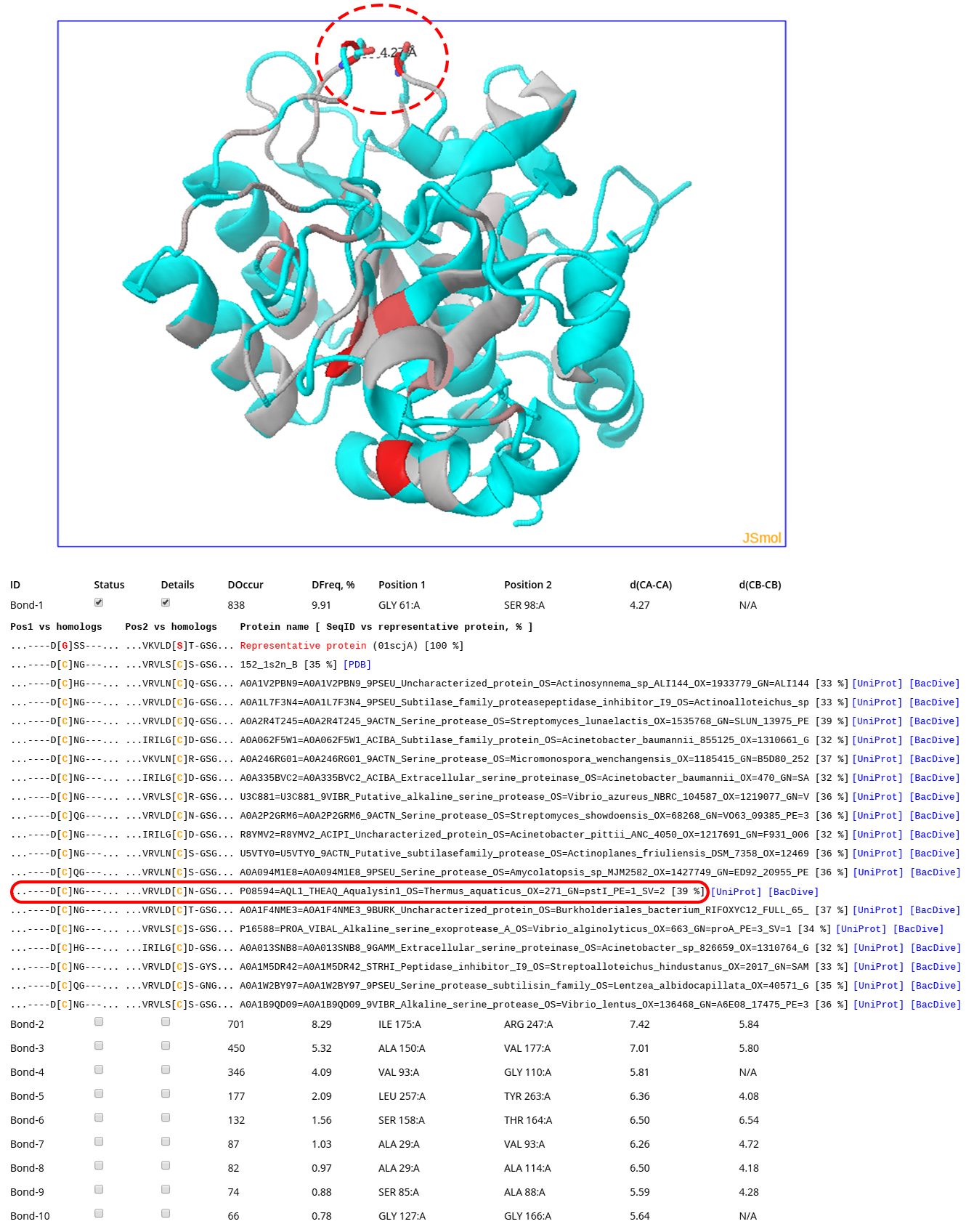 Click here to enlagre
Click here to enlagre
The Analysis section implements HTML5-based interactivity to study Yosshi output on-line. The information presented at the on-line analysis page is equivalent to the PyMol PSE structural annotation file and the text summary of the annotation, which are discussed above. The screenshot above corresponds to the analysis of subtilisin E from Bacillus subtilis and the subtilisin-like serine proteases superfamily, which can be reviewed by activating the “Demo mode” at the Yosshi submission page (this example is discussed in details here). In brief, The structure of subtilisin E from Bacillus subtilis is shown in the 3D viewer. The backbone of the protein is gradient painted from grey to red according to the expected abundance of an S-S bond at the corresponding positions in structures of homologs, with intensive red indicating pairs of positions most frequently hosting disulfides. A pair of residues Gly61 and Ser98 has a rank #1 in the Yosshi output (shown as sticks and indicated by a dashed oval) as 838 proteins were discovered to contain cysteines in equivalent positions. The corresponding sub-sequences of some of these homologs are shown (cysteines are colored in yellow). The entry corresponding to a homolog from a thermophilic organism (Aqualysin-1 from Thermus aquaticus) is indicated by a red oval. For selected proteins the HTML links to respective pages in PDB/UniProt/BacDive databases are highlighted in blue (click here to learn more about this feature).