Yosshi protocol
The Yosshi workflow consists of bioinformatic analysis to search for pairs of cysteine residues in sequences of homologs, and structural filtration to evaluate whether introduction of the selected cysteines at corresponding positions in the user-submitted query protein can result in a formation of a disulfide bond. The outline of the process is briefly described below. The Yosshi algorithm is discussed in details in the corresponding publication.
Outline of the Yosshi algorithm
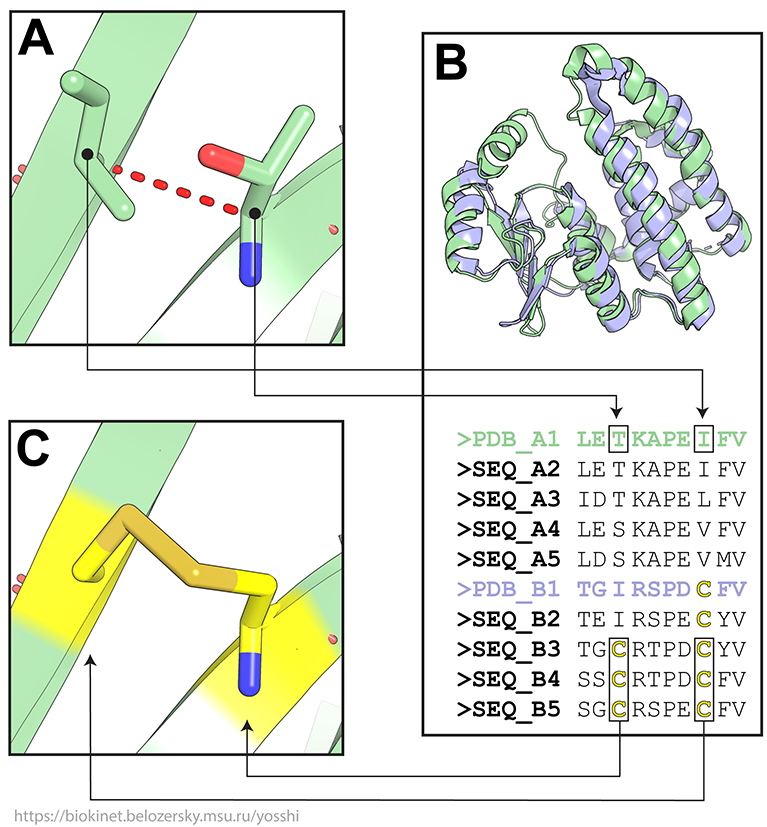
(A) The first tier filter is applied to select all pairs of amino acid residues which are located close enough in the query structure for further inspection;
(B) This initial set of pairs of positions is further subjected to bioinformatic analysis of the available sequence and structural data of the homologous proteins within a superfamily to select only such pairs which are both occupied by cysteine residues in at least one homolog of the query protein;
(C) Finally, the 3D-motif analysis is implemented to evaluate whether the selected pairs of positions can form a disulfide bond assuming the respective amino acids are mutated to cysteines in the query protein structure, and prepare 3D-models of corresponding mutants.
The key part of the Yosshi workflow is the bioinformatic analysis what requires a large set of proteins expected to have diverse properties and different disulfide connectivity patterns within a common structural core of a superfamily to be collected and aligned, representing methodological and computational challenges. This step is facilitated by the integrated Mustguseal web-server capable of constructing large structure-guided sequence alignments of functionally diverse protein families that can include thousands of proteins automatically. A detailed guideline on automatic preparation of the input data within the Yosshi+Mustguseal workflow is provided here.