Yosshi
A web-server for disulfide engineering by bioinformatic analysis of diverse protein families
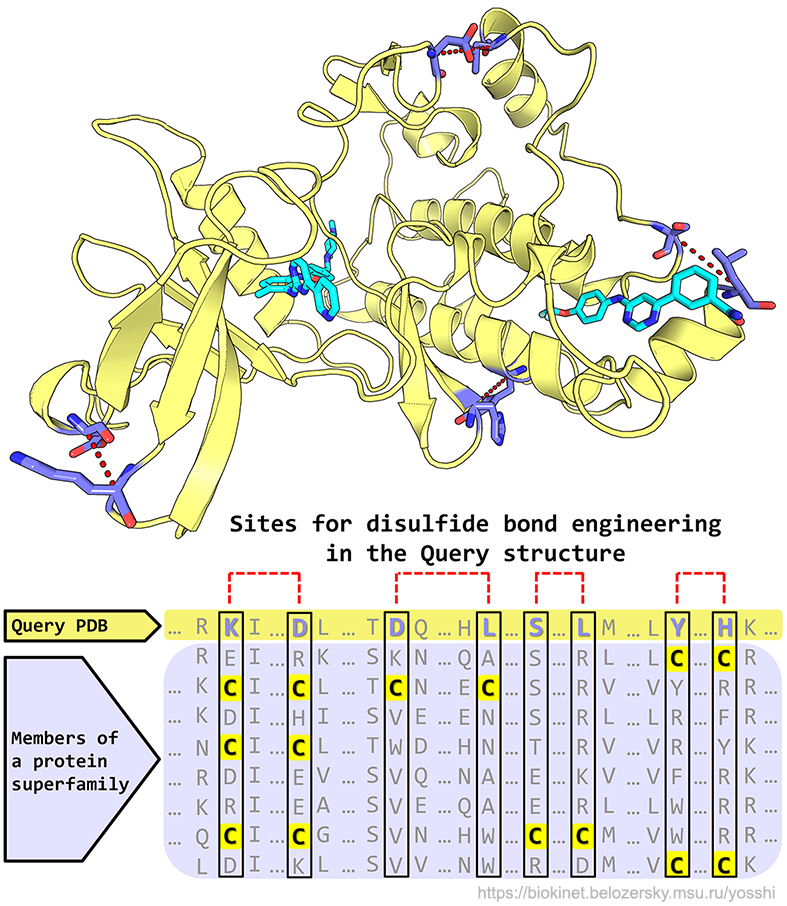

... to systematically classify and study disulfide bonds in diverse protein families,
and to assist at selecting hot-spots for disulfide engineering in the structure of your query protein
Version 1.1 since October 31st, 2019
THIS WEB-SITE/WEB-SERVER/SOFTWARE IS PROVIDED “AS IS”, WITHOUT WARRANTY OF ANY KIND, EXPRESS OR IMPLIED, INCLUDING BUT NOT LIMITED TO THE WARRANTIES OF MERCHANTABILITY, FITNESS FOR A PARTICULAR PURPOSE AND NONINFRINGEMENT. IN NO EVENT SHALL THE AUTHORS OR COPYRIGHT HOLDERS OR HARDWARE OWNERS OR WEB-SITE/WEB-SERVER/SOFTWARE MAINTEINERS/ADMINISTRATORS BE LIABLE FOR ANY CLAIM, DAMAGES OR OTHER LIABILITY, WHETHER IN AN ACTION OF CONTRACT, TORT OR OTHERWISE, ARISING FROM, OUT OF OR IN CONNECTION WITH THE WEB-SITE/WEB-SERVER/SOFTWARE OR THE USE OR OTHER DEALINGS IN THE WEB-SITE/WEB-SERVER/SOFTWARE. PLEASE NOTE THAT OUR WEBSITE COLLECTS STANDARD APACHE2 LOGS, INCLUDING DATES, TIMES, IP ADDRESSES, AND SPECIFIC WEB ADDRESSES ACCESSED. THIS DATA IS STORED FOR APPROXIMATELY ONE YEAR FOR SECURITY AND ANALYTICAL PURPOSES. YOUR PRIVACY IS IMPORTANT TO US, AND WE USE THIS INFORMATION SOLELY FOR WEBSITE IMPROVEMENT AND PROTECTION AGAINST POTENTIAL THREATS. BY USING OUR SITE, YOU CONSENT TO THIS DATA COLLECTION.
Publication: Suplatov D.A., Timonina D.S., Sharapova Y.A., Švedas V.K. (2019) Yosshi: a web-server for disulfide engineering by bioinformatic analysis of diverse protein families. Nucleic Acids Res., 47(W1), W308–W314. DOI:10.1093/nar/gkz385 Conference: Suplatov D.A., Timonina D.S., Sharapova Y.A., Švedas V.K. (2019) Yosshi: the bioinformatic approach to protein disulfide engineering. MCCMB'2019, poster: [download]
Navigation:
- Prerequisites and compatibility
- The Yosshi protocol
- Preparing the input data for Yosshi
- Preparing the input data for multiple-chain proteins
- The Yosshi parameters
- The Yosshi output
- The Yosshi example
- The Yosshi file sharing and security features
- Citing Yosshi
Highlights:
- The "YOur web-server for S-S bond HarvestIng" is a new highly automated on-line tool for a systematic homology-driven analysis and engineering of disulfide bonds that can be easily used by a general biologist at a daily laboratory routine;
- The Yosshi facilitates a broader interpretation of disulfides not just as a factor of structural stability, but rather as a mechanism to implement diversity within a superfamily;
- The key novelty of Yosshi is implementation of the bioinformatic analysis to search for pairs of cysteine residues in sequences of homologs followed by the 3D-motif analysis to evaluate whether introduction of the selected cysteines at corresponding positions in the user-submitted query protein can result in a formation of a disulfide bond, as outlined here;
- The bioinformatic analysis is supported by the integrated Mustguseal web-server to construct large structure-guided sequence alignments of functionally diverse protein families;
- The output of Yosshi is a detailed homology-based annotation of disulfides within a common structural fold of a superfamily. Disulfides conserved in larger groups of proteins are shown first suggesting an important role in a common function or property shared by these homologs;
- The Yosshi+Mustguseal integrated web-based method provides an intuitive and easy-to-use interface to study the abundance of S-S bonds within a superfamily, compare disulfide connectivity in homologs with different properties, as well as to identify disulfide bridges present in homologs but not in the query protein that can be introduced to design its stability and functional properties.